Opportunities for different HBO/BSc/MSc thesis projects
There are several opportunities for student projects related to my different research projects that can be carried out in the Biosystematics group at WUR often co-supervised by my colleagues and/or PhD students. If you are interested in one of the below offered projects, you can contact me by email to discuss a thesis possibility on the topic of your interest. As a WUR student, you can also visit the TIP-WUR database to get idea of other project connected to Biosystematics.
Natural variation of butterfly egg-killing cell death in different Brassica populations
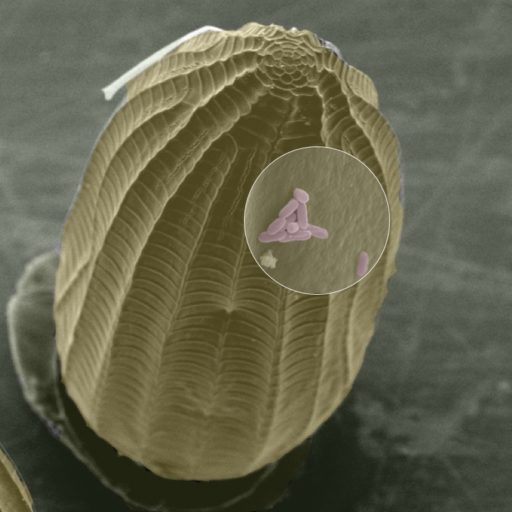
Eggs deposited by cabbage white butterfly eggs (Pieris spp) induce a so-called hypersensitive response (HR)-like cell death in brassicaceous plants, including cabbage crops. This response can lead to the killing of the eggs and is thus a unique and underexplored first line-of-defense trait that helps plants to get rid of enemies before they become destructive. Programmed cell death or HR was originally described as a defense response to plant pathogens. The model plant Arabidopsis thaliana activates a response that is similar to the recognition of pathogen-associated molecular patterns after oviposition.
The aim of this project is to study the genetic mechanism of this HR-like response in different populations of the black mustard (Brassica nigra). Interestingly, some genotypes of this plant express a very strong necrosis against eggs, while others dont and we want to understand the underlying mechanism of this phenotypic variation therein.
You will:
- explore natural variation in this plant trait in different European populations (Sweden, NL, South France) of B. nigra
- perform Pieris egg survival tests on plants from the different populations
- perform transcriptional analysis of resistance genes
Skill developmet:
- phenotyping, qPCR analysis, bioassays with insects, optional: field work in NL, Sweden and/or South France
Requirements:
- Basic knowledge of insect and plant ecology, molecular biology and bioinformatics
Natural variation in elicitation of butterfly egg-killing cell death in different Pieris population
Eggs deposited by cabbage white butterfly eggs (Pieris spp) induce a so-called hypersensitive response (HR)-like cell death in Brassica plants, including B. nigra. This response can lead to the killing of the eggs and is thus a unique and underexplored first line-of-defense trait that helps plants to get rid of enemies before they become destructive. We want to understand which molecules from the egg and egg glue, i.e. egg-associated molecular patterns (EAMP) are detected by the plant. Is there natural variation in the elicitation of this necrosis on the butterfly side; in other words, do some butterflies avoid that their eggs are detected by the plant.
The aim of this project is to study natural variation in elicitation of HR in different populations of Pieris. To understand the biosynthesis of the EAMP, we want to find the genes underlying the biosynthesis. One way to do so, is to find natural variation and performe a QTL mapping by using strains that differen in in the trait and cross them to build up a mapping population.
You will:
- explore natural variation in induction of HR in B. nigra by eggs of Pieris butterflies from different European populations (Sweden, NL, South France)
- Perform a Mendelian crossing scheme: cross parental strains that differ in the induction of HR, phenotyp the F1 population and backcross them with the inducing and non-inducing parent
Skill developmet:
- phenotyping, classical genetics, optional: field work in NL, Sweden and/or South France
Requirements:
- Basic knowledge of insect and plant genetics, ecology
Transciptional analysis of genes involved in elicitation of butterfly egg-killing cell death
Eggs by cabbage white butterfly eggs (Pieris spp) induce a so-called hypersensitive response (HR)-like cell death in Brassica plants, including B. nigra. This response can lead to the killing of the eggs and is thus a unique and underexplored first line-of-defense trait that helps plants to get rid of enemies before they become destructive. We want to understand which molecules from the egg and egg glue, i.e. egg-associated molecular patterns (EAMPs), are detected by the plant. We know that the EAMPs are located in the accessory reproductive glands (ARGs) of female butterflies that produce the egg glue. The butterfly uses the egg glue to attach the eggs onto the leaf.
The aim of this project is to perform a transcritional analysis of the ARGs to find genes involved in the biosynthesis of the EAMPs.
You will:
- dissect ARGs of Pieris butterflies from different European populations (Sweden, NL, South France)
- Perform an RNA-seq from the ARGs of different
- Analyse sequenced data using a bioinformatics pipeline
- quantify expression of candidate genes
Skill developmet:
- Molecular biology techniques (RNA isolation, nanopore sequencing, qPCR), bioinformatics, insect dissection
Requirements:
- Basic/good knowledge of molecular biology, bioinformatics, insect ecology
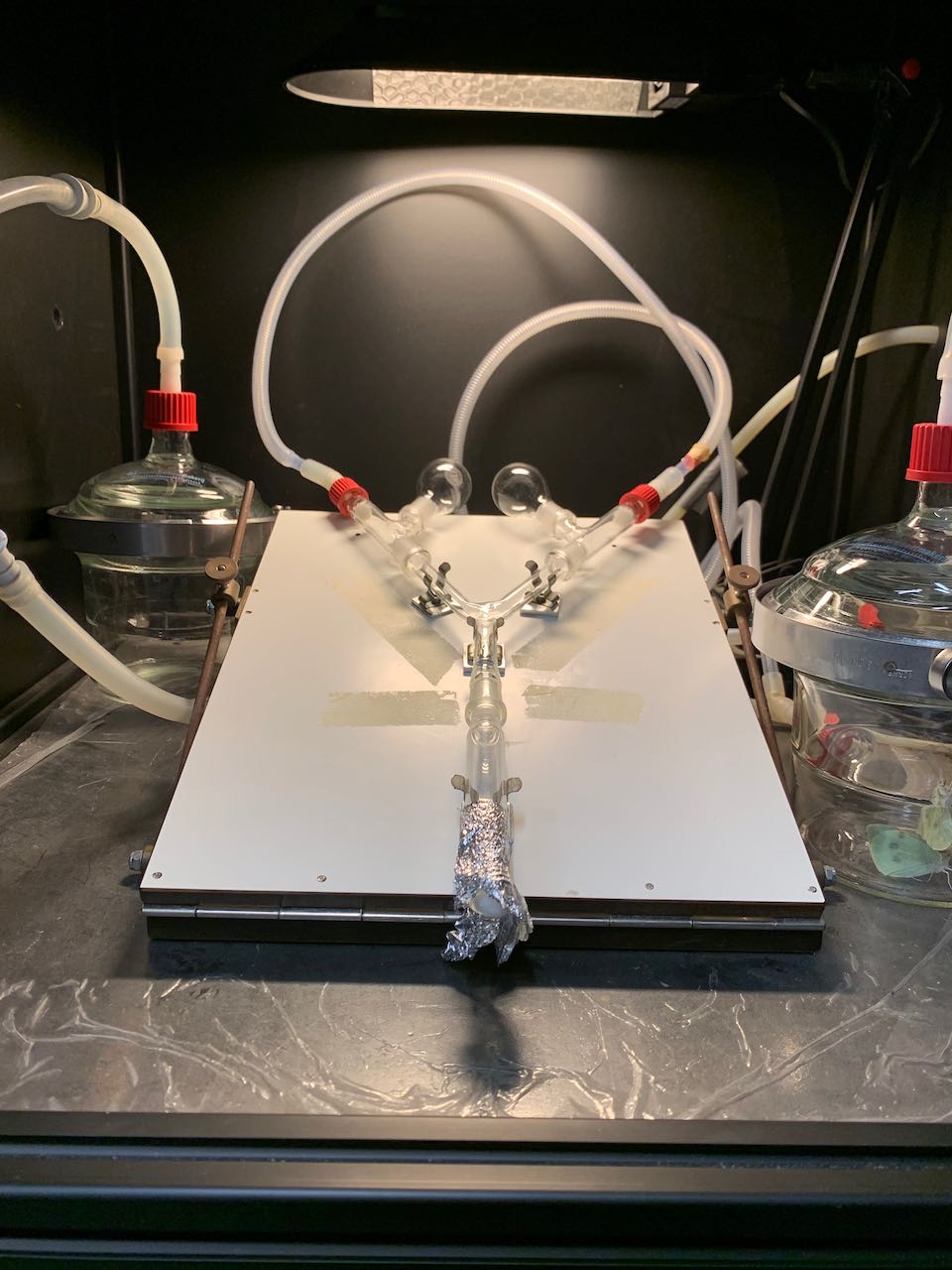
Exploring the responses of Trichogramma wasps to oviposition-induced plant volatiles
Several species of the minute egg parasitoids in the genus Trichogramma have been found to parasitise the eggs of cabbage white butterflies (Pieris) and are now used in their biological control. One way that these wasps find butterfly eggs to parasitise is by using plant volatiles induced by insect oviposition (oviposition-induced plant volatiles - OIPVs). In this project, your aim will be to further elucidate the prevalence and specificity of Trichogramma attraction to Pieris OIPVs.
You will:
Do field surveys and behaviour assays to investigate prevalence and specificity, rear butterflies and collected wasps for behavior experiments, use identify wasps’ species and haplotype, design and conduct a series of behavioural assays to determine the specificity of attraction to different wasp, plant and butterfly combinations.
Skill development:
- Various olfactometers tests: Y-tube, T-maze, etc
- DNA barcoding
- phylogenetic network analysis using the genetic data to trace newly determined behavioral traits to analyse evolutionary patterns and host specificity.
Requirements:
- Basic/good knowledge of molecular biology, insect ecology and behavior
Comparing and linking butterfly pheromones and parasitism across Europe
Is there intra-specific variation in the pheromones of Pieris butterflies, and/or in the selection pressure they face from hitchhiking egg parasitoids?
Three species of cabbage white butterflies (Pieris) are known to use an anti-aphrodisiac pheromone in their sexual communication. Several species of the minute egg parasitoids in the genus Trichogramma have been found to use these pheromones as cues to find mated females to climb onto in order to hitchhike with them to freshly laid eggs. If this phenomenon is widespread and mortality posed to butterflies is hight, we expect natural selection on these pheromones to possibly constrain the use and evolution of the AA, or possibly exert pressure for changes to use different compounds or blends. To investigate this, we will compare pheromones of these butterflies in populations where there is variation in the selection pressure posed by hitchhiking parasitoids. You will determine whether AAs and/or parasitism vary between these sites, and thus investigate a possible link.
You will:
- Continue field surveys of hitchhiking egg parasitoids in Wageningen
- Conduct field work at a field site in Lund, Sweden, where you will collect and rear the local butterflies and parasitoids
- Identify egg parasitoids and compare host-parasite networks between sites
- Collect volatiles from butterflies of the different populations to analyse their AAs with GC-MS
Skill development:
- GC/MS analysis, DNA barcoding, field work, constructing phylogenetic trees
Requirements:
- Basic/good knowledge of molecular biology, insect ecology and behavior